Biological sciences
March 23, 2022
How a particular protein regulates up to two-thirds of the world's methane emission: key mechanism for methane emission in anaerobic environments is now understood, potentially helping the fight against climate change
Two-thirds of global methane release is likely due to natural emission during anaerobic (oxygen-free) activity of single-celled microorganisms called archaea. The protein mechanism has now been understood using biophysical and biochemical techniques.
While methane accounts for about 16% of the abundance in the atmosphere of greenhouse gases - which also include carbon dioxide, nitrous oxide, water vapor – it is more than 25 times better than carbon dioxide at trapping heat. Two-thirds of global methane release is believed to be through natural emission during anaerobic activity of primitive single-celled microorganisms called archaea. Understanding the precise mechanism by which archaea produce methane could lead to technology that reduces methane production by archaea and helps in the fight against global warming.
Archaea are distinct from bacteria mainly because of their habitat and sources of energy. The so-called methanogen archaea emit methane as a byproduct of energy generation necessary for their survival. The biomolecule responsible for methane formation is the so-called Methyl-Coenzyme M Reductase (or MCR) protein that induces the chemical conversion. In order for MCR to catalyze this reversible reaction, it needs to be activated by a partner protein that belongs to the superfamily of B12-dependent radical S-Adenosyl-L-Methionine (or SAM) enzymes.
The superfamily of radical SAM enzymes contains over 200,000 independent-sequenced proteins. It has been associated with a multitude of natural processes, including the biosynthesis of antibiotics and chlorophyll. One of these key enzymes (Mmp10) is responsible for the activation of the MCR protein and is therefore involved in the regulation of its methane formation. The ubiquity of SAM enzymes across the biosphere reflects their importance in catalyzing reactions that are fundamental to all types of life. However, the mechanisms balancing their biological activities remains poorly understood.
To decipher the activities of the Mmp10 SAM enzyme, Dr. Olivier Berteau, from the Micalis Institute, Université Paris-Saclay, assembled a team of scientific experts with various complementary areas of expertise, including other researchers from that university, Aix Marseille University and Synchrotron SOLEIL in France, as well as Nagoya University in Japan. The results of the investigation were published online in the journal Nature on Feb 2, 2022.
Key to the activity of B12-dependent radical SAM enzymes is a simple yet powerful mechanism for triggering the catalytic reaction. The difficulty in getting the enzyme to simultaneously accommodate all the actors involved in the reaction has meant that little structural information had been available that could help explain how the reaction works.
To remedy this, the research team combined crystallographic results with biochemical and biophysical data to explain how B12-dependent radical SAM proteins regulate their activity, down to atomic-level details. The Mmp10 enzymatic mechanism was imaged with all actors of the reaction present.
The results of this research have implications for the development of biotechnologies that would control key enzymatic events, particularly those implicated in methane emission, helping in the fight against global warming.
Co-author Professor Leo Chavas, of Nagoya University, is excited by the results of this long-term investigation. “A total of 137 proteins were screened at a leading synchrotron facility in France to get a glimpse of these rare events, which are so difficult to catch. This research also opens the door to biotech developments.”
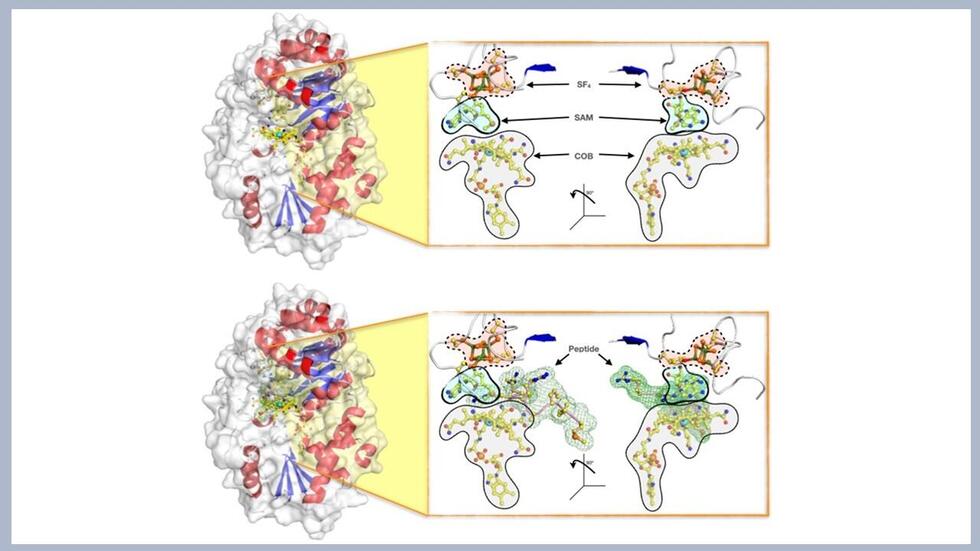
Surface illustrations of the enzyme Mmp10, showing (top) the protein with the complete set of co-factors required for the enzymatic reaction to occur (namely: the SF4 cluster, the SAM and the cobalamin COB), and (bottome) the enzyme in action while accommodating the peptide for Methyl transfer. The research team has elucidated the crystallographic details of the reaction and proposed a mechanism by which the protein activates a partner protein that stands as a major contributor of naturally occurring methane production in archaea (Credit: Leonard Chavas)
The paper, "Crystallographic snapshots of a B12-dependent radical SAM methyltransferase," was published in the journal Nature, Feb 2, 2022 and is available at doi.org/10.1038/s41586-021-04355-9.
Authors:
Cameron D. Fyfe, Noelia Bernardo-García, Laura Fradale, Alain Guillot, Clémence Brewee, Alhosna Benjdia, Olivier Berteau (Micalis Institute, France); Stéphane Grimaldi (Aix-Marseille University, France); Pierre Legrand (Synchrotron SOLEIL, France); Leonard Michel Gabriel Chavas (Nagoya University, Japan).
Contact:
Leonard Chavas
Professor, Synchrotron Radiation Research Center and Department of Applied Physics, Graduate School of Engineering, Nagoya University
E-mail: l.chavas@nusr.nagoya-u.ac.jp
Funding:
This work was supported by the European Research Council (ERC consolidator grant 617053), ANR (ANR-17-CE11-0014) and the CHARMMMAT Laboratory of Excellence (ANR-11-LABX0039).